Module 3 Self Assessment
Things Learnt:
Technical:
- Generating Quality Control reports
- Identifying some outliers from the QC stats plot
- Performing normalisation and background correction using RMA
- Generating RLE and NUSE boxplots and summary statistics
- Learning the purpose behind performing quality control and data pre-processing
- Learning more about data visualisation using PCA and heatmaps
Tools:
- R studio / R cloud
- Packages- simpleaffy, arrayQualityMetrics, affyQCReport, affyPLM
- Github
Soft Skills:
Time management, Virtual Collaboration with my group members, working with other team members and helping each other out when we face certain problems or roadblocks, collectively coming up with solutions for them.
Achievements:
- Tried more than one method of performing quality control and normalisation
- Interpreted and presented our results
- Identified some outlier values by comparing samples using the geo2r table
Difficulties Faced: affyQCreport takes a lot of time and memory to run and Rstudio kept crashing when I tried to run it on my laptop
Module 4 Self Assessment
Things Learnt:
Technical:
- Removing outliers from data
- Gene annotation and gene filtering
- Limma Analysis
- Generating a volcano plot
- Other methods of visualising the top differentially expressed genes (DEGs)
- Analysing selected samples in geo2r
Rshiny app: This week I also worked on designing a general layout for grouping of samples along with Aditi in group B2 using Figma.
Tools:
- geo2r
- EnhancedVolcano
- Limma
- hgu133plus2.db
Soft Skills:
- Collaborating with my team member Leila this week
- Asking other groups for help and learning from their mistakes while troubleshooting, comparing my code and results with theirs
- Presentation skills
Achievements:
- Successfully performed gene annotation and gene filtering
- Successfully generated a table of top DEGs and volcano plot that matched with geo2r values
Difficulties Faced:
- I initially faced some difficulty in creating the desired design matrix and contrast matrix
- After that I was able to generate a volcano plot with only the probe ids and had some trouble with the gene annotation. However other team members like Leila, Ivan and Arian helped me out a lot with that part!
Module 6 Self Assessment
Things learnt:
Technical/Tools:
- Performed pathway enrichment using 3 web based tools–Enrichr, DAVID and Metascape
- Explored other functional analyses using these tools as well
- Also explored results using Reactome and Wikipathways besides KEGG
Soft Skills: Collaborating with my teammate Shreya this week, Time management, Presenting our findings
Achievement Highlights:
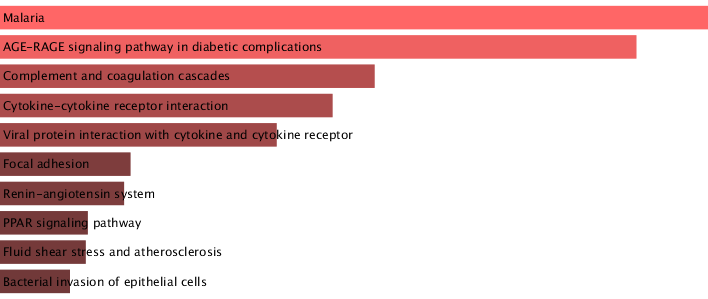
- Successfully generated results that matched our results from module 5, where Malaria and AGE-RAGE signalling pathway were highlighted as top enriched KEGG pathways
Results using Enrichr:
Difficulties faced:
- Initially, Shreya and I faced some difficulty using DAVID and how to visualise our data on it